Overview
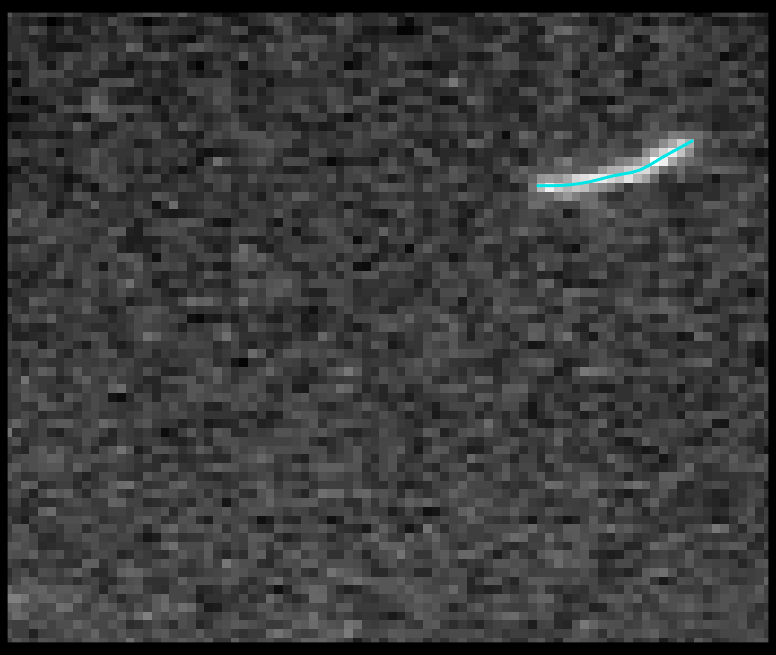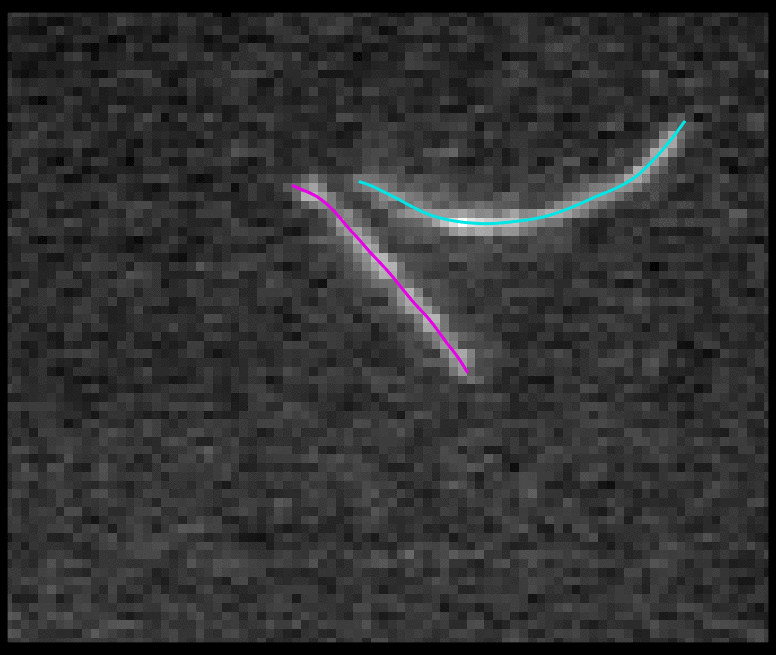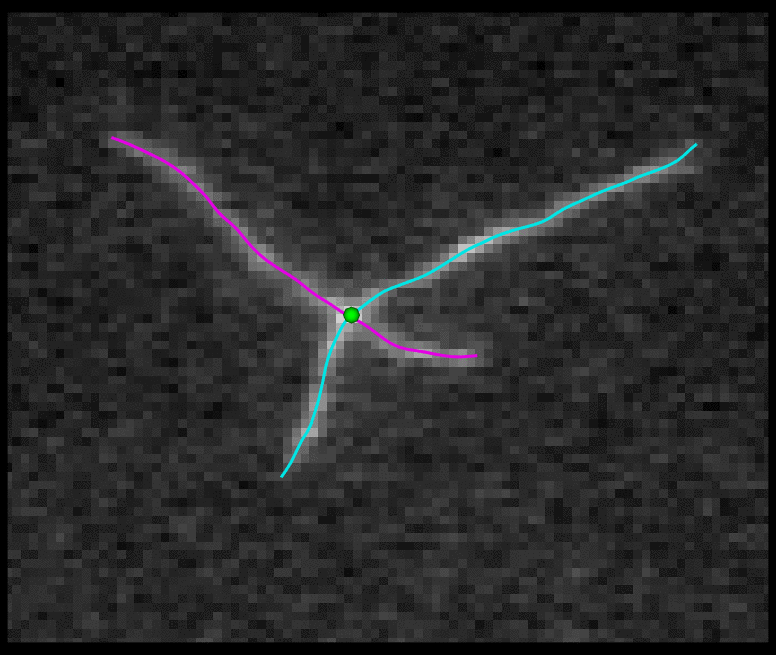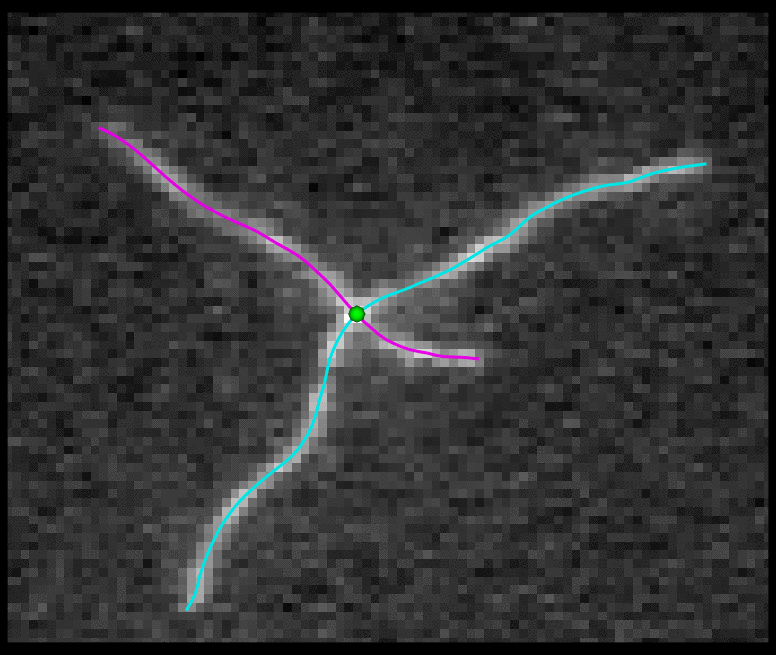
TSOAX is an open source software to extract and track the growth and deformation of biopolymer networks from 2D and 3D time-lapse sequences. It tracks each filament or network branch from complex network dynamics and works well even if filaments disappear or reappear. The output is a set of tracks for each evolving filament or network segment.
TSOAX is an extension of SOAX (for network extraction in static images) to network extraction and tracking in time lapse movies.
TSOAX facilitates quantitative analysis of network dynamics of multi-dimensional biopolymer networks imaged by various microscopic imaging modalities. The underlying methods of TSOAX includes multiple Stretching Open Active Contour Models for extraction and a combined local and global graph matching framework to establish temporal correspondence among all extracted structures. TSOAX is built with a cross-platform Qt GUI with 3D interactive visualization for exploring image sequences and visually checking results. It also provides utilities such as image resampling, taking screen snapshots, etc.
Reference
T. Xu, C. Langouras, M. Adeli Koudehi, B. Vos, N. Wang, G. H. Koenderink, X. Huang and D. Vavylonis, "Automated Tracking of Biopolymer Growth and Network Deformation with TSOAX": Scientific Reports 9:1717 (2019)
Credits
TSOAX was designed and implemented by Ting Xu advised by Professor Xiaolei Huang and Professor Dimitrios Vavylonis at Lehigh University, in collaboration with the co-authors of the reference above. Further improvements were provided by David Rutkowski at Lehigh University.
This work is supported by NIH grants R35GM136372, R01GM114201 and R01GM098430.
