My research group is investigating the molecules and mechanisms that maintain chromosomes in order to control cell viability and proliferation. In particular, we study the enzyme telomerase, which consists of RNA and protein components and is centrally important to human cancer and aging, and how it prevents the erosion of telomeric DNA at chromosome ends during cell division.
As briefly outlined below, we are discovering and investigating (1) essential telomerase RNA functional elements and their mechanisms of action, (2) the molecular mechanisms that maintain telomere length, and (3) the molecular and cellular hallmarks of senescence caused by critically short telomeres.
The telomerase ribonucleoprotein (RNP) enzyme, telomeres, and senescence.
What is the physical organization of telomerase RNA and how does it relate to function?
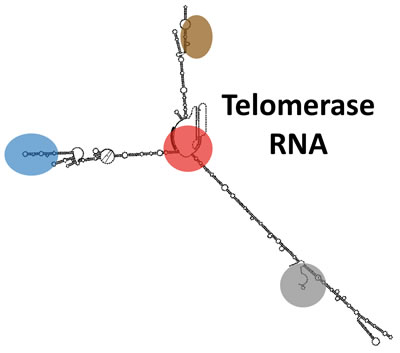
I determined the secondary structure of the 1157-nt S. cerevisiae telomerase RNA while I was a postdoc and revealed that it functions as a flexible scaffold for the essential Est1 protein subunit. With NIH K99/R00 funding, my independent lab at Johns Hopkins University then demonstrated that this novel function for an RNA in a ribonucleoprotein complex also pertains to the other two S. cerevisiae telomerase holoenzyme-specific subunits, Ku and Sm7, as well as for the essential three-way junction in the evolutionarily distant fission-yeast telomerase RNA.
My lab has also discovered several essential well-conserved RNA structural features within yeast and human telomerase RNAs. The characteristics of some of the new elements demonstrate that telomerase RNAs also have functions beyond acting as a flexible scaffold for protein subunits.
We are now determining the binding interface between the RNA and the catalytic protein subunit (TERT) at the core of the RNP enzyme, the extent of flexibility of telomerase RNA in the holoenzyme, as well as new telomerase subunits and their functions.
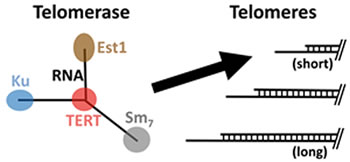
Telomerase is preferentially recruited to short telomeres to promote their extension, but how this occurs is not known. We recently discovered how telomerase is recruited to telomeres by its Ku subunit. This finding led us to a model for the molecular mechanism of telomere-length homeostasis in yeast, as well as why there are two telomerase-recruitment pathways, coordinated by the Ku and Est1 subunits. With an NIH R01 grant supporting this project, we are now testing our model for the molecular mechanism coordinating telomere-length homeostasis. One aim is to determine the arrangement of telomere proteins along chromosome ends to help learn how they “sense” telomere length to preferentially recruit telomerase to short telomeres.
What are the molecular and cellular hallmarks of senescence caused by telomere loss?
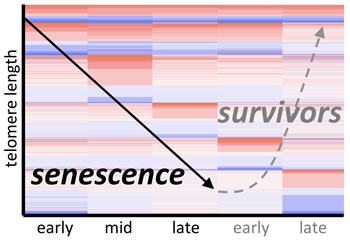
Using RNA-seq, we have determined the genome-wide transcriptional response of cells undergoing senescence due to erosion of telomeres, as well as the “survivors” that emerge subsequently. Several major findings have come from these RNA-seq data: (1) senescence has distinct phases, each with a signature of hundreds of differentially expressed genes; (2) senescence is characterized by upregulation of >100 novel lncRNAs; and (3) there are many induced cellular pathways. We are now actively identifying the roles of differentially expressed proteins and RNAs to define and characterize the hallmarks of cellular senescence caused by telomere erosion.
Mefford, M.A., Hass, E.P., and Zappulla, D.C. (2020) A 4-base pair core-enclosing helix in telomerase RNA is essential and binds to the TERT catalytic protein subunit. bioRxiv doi: https://doi.org/10.1101/2020.01.10.902601
Hass, E.P. and Zappulla, D.C. (2017) A yeast two-hybrid system based on CRISPR-dCas9 for investigating RNA-protein interactions. bioRxiv doi: https://doi.org/10.1101/139600
Hass, E.P. and Zappulla, D.C. (2020) Repositioning the Sm-binding site in S. cerevisiae telomerase RNA reveals RNP organizational flexibility and Sm-directed 3’-end formation.
(in press)Chen, H., Xue, J., Churikov, D., Hass, E.P., Lemon, L.D., Luciano, P., Bertuch, A.A., Zappulla, D.C., Geli, V., Wu, J., and Lei.M. (2018) Structural insights into yeast telomerase recruitment to telomeres. Cell 172:331–343.
*Johns Hopkins University School of Medicine press release
Niederer, R.O., Papadopoulos, N. and Zappulla, D.C. (2016) Identification of novel noncoding transcripts in telomerase-negative yeast using RNA-seq. Scientific Reports 6, 19376; doi: https://doi.org/10.1038/srep19376
Mefford, M.A. and Zappulla, D.C. (2016) Physical connectivity mapping by circular permutation of human telomerase RNAreveals new regions critical for activity and processivity. Molecular and Cellular Biology 36(2):251–261.
Hass, E.P. and Zappulla, D.C. (2015) The Ku subunit of telomerase binds Sir4 torecruit telomerase to lengthen telomeres in S. cerevisiae. eLIFE 4:e07750.
*Highlighted on journal cover
*Johns Hopkins University press release: http://hub.jhu.edu/2015/09/14/telomere-proteins-cancer-aging
Lebo, K.J., Niederer, R.O. and Zappulla, D.C. (2015) A second essential function of the Est1 arm of yeast telomerase RNA. RNA 21:862–876.
Niederer, R.O., and Zappulla, D.C. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA 21:254–261.
Mefford, M.A., Rafiq, Q., and Zappulla D.C. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. (2013) EMBO Journal 13:32(22): 2980–2993.
Lebo K.J. and Zappulla D.C. Stiffened yeast telomerase RNA supports function in vitro and in vivo. (2012) RNA 18: 1666–1678.
Zappulla D.C., Goodrich K.J., Arthur J.R., Gurski L.A., Denham E.M., Stellwagen A.E. and Cech T.R. (2011) Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA 17:298–311. (corresponding author)
Zappulla D.C.*, Roberts J.N., Goodrich K., Cech T.R. and Wuttke D.S.* (2009) Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p. (*co-corresponding author) Nucleic Acids Research 37:354–367.
Box J.A., Bunch J.T., Zappulla, D.C., Glynn E.F., and Baumann P. (2008) A flexible template boundary element in the RNA subunit of fission yeast telomerase. Journal of Biological Chemistry 283(35):24224-33
Zappulla D.C. and Cech, T.R. (2006) RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harbor Symposia on Quantitative Biology (Symposium 71: Regulatory RNAs) 71:217–224.
Zappulla D.C., Maharaj A.M., Connelly J., Jockusch R., and Sternglanz R. (2006) Rtt107/Esc4 binds silent chromatin and DNA repair proteins using different BRCT motifs. BMC Molecular Biology 4: 40-72.
Zappulla D.C., Goodrich K., and Cech T.R. (2005) A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nature Structural and Molecular Biology 12(12):1072-1077.
Zappulla D.C. and Cech, T.R. (2004) Yeast telomerase RNA: a flexible scaffold for protein subunits. Proceedings of the National Academy of Sciences101(27): 10024-10029.
Andrulis E.D., Zappulla D.C., Alexieva-Botcheva, K., Evangelista, C. and Sternglanz, R. (2004) Targeted silencing screens at HMR identify novel transcriptional silencing factors. Genetics 166:631-635.
Zappulla D.C., Sternglanz, R., and Leatherwood, J. (2002) Control of DNA replication timing by a transcriptional silencer. Current Biology 12: 869-875.
Andrulis E.D.*, Zappulla D.C.*, Ansari A.*, Perrod S., Laiosa C.V., Gartenberg M.R., and Sternglanz, R. (*co-first authors) (2002) Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Molecular and Cellular Biology 22(23): 8292-8301.
Xie W., Gai X., Zhu Y., Zappulla D.C., Sternglanz R., and Voytas, D. (2001) Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Molecular and Cellular Biology
Andrulis E.D., Neiman A.M., Zappulla D.C., and Sternglanz R. (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592-595.
Ong B.C., Zimmerman A.A., Zappulla D.C., Neufeld E.J., and Burrows F.A. (1998) Prevalence of factor VLeiden in a population of patients with congenital heart disease. Canadian Journal of Anesthesia 45(12): 1176-1180.
Tufarelli C., Fujiwara Y., Zappulla D.C., and Neufeld E.J. (1998) Hair defects and pup loss in mice with targeted deletion of the first cut repeat domain of the Cux/CDP homeoprotein gene. Developmental Biology 200(1): 69-81.
Yandava C.N., Zappulla D.C., Korf B.R., and Neufeld E.J. (1996) ARMS test for diagnosis of factor VLeiden mutation, a common cause of inherited thrombotic tendency. Journal of Clinical Laboratory Analysis 10(6): 414-417.
The Zappulla Lab is currently searching to fill the following positions:
- Postdoctoral Research Associate
- Graduate Students
Contact Dr. Zappulla at dcz218 [at] lehigh.edu or 610-758-5088
Current lab members:
Ariana Amendolara (PhD student)
Nick Cann (rotating PhD student)
Allison Peeney (rotating PhD student)
Kayli Silimperi (PhD student)
Anna Nicosia (undergraduate researcher)
Chaihong Nah (undergraduate researcher)
Zappulla Lab alumni:
Dr. Kevin Lebo (PhD student) — postdoc with Ron Weiss (MIT), now Scientist at Ring Therapeutics (Cambridge, MA)
Dr. Rachel Niederer (PhD student) — now postdoc with Dr. Wendy Gilbert (Yale University)
Dr. Karen McMurdie (PhD student) — now Scientist II at BD Biosciences (Sparks, MD)
Dr. Evan Hass (PhD student) — now postdoc with Dr. John Rinn (HHMI, University of Colorado — Boulder)
Dr. Melissa Mefford (postdoc) — now tenure-track faculty at Morehead State University


