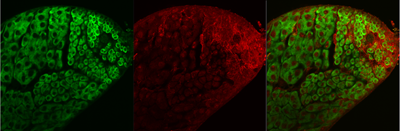
The Ware lab studies components of one of the most abundant and complex machines in all cells – the ribosome.
This complex of several ribosomal RNAs and multiple ribosomal proteins is the macromolecular “stage” on which all proteins are synthesized in cells in a process called translation. As an essential complex in all cells, the ribosome varies in composition and structure between different organisms, prompting questions of how those structural differences may affect translation or the regulation of translation in different types of cells.
Using biochemical, cellular, genetic, and molecular approaches, the Ware lab is studying differences in ribosome composition and biogenesis at the RNA and protein levels in the fruit fly, Drosophila melanogaster. We are studying the eukaryotic-specific eRpL22 ribosomal protein family where duplicate genes encoding eRpL22 and eRpL22-like (called paralogues) are present to determine if paralogue roles within the ribosome are redundant and to determine if paralogues specify novel, extra-ribosomal roles. Functional differences may have evolved over time, allowing for the development of new or refined functions for these ribosomal protein paralogues. Tissue-specific expression of eRpL22 paralogues in the fly testis and developing eye has provided evidence that paralogue functions are not completely redundant. Our recent investigations support a role in translation regulation for paralogue-specific ribosomes within the male germline, demonstrating that subsets of mRNAs and noncoding RNAs are differentially-enriched on polysomes containing either eRpL22 or eRpL22-like. We propose that ribosome heterogeneity (here illustrated by assembly of different eRpL22 paralogues into ribosomes) defines “specialized ribosomes” with a role in regulating translation of different mRNAs during spermatogenesis. The mechanism for RNA selection on specific ribosome types is currently under investigation.
|
Ware lab research focuses on phage biology and functional genomics
Phage biology research in the Ware lab was spawned from research in Lehigh University’s SEA-PHAGES Program (sponsored by HHMI). Beyond the SEA-PHAGES goals of discovery of novel actinobacteriophages and phage genome annotation, the Ware lab focuses on characterization of novel phage genes with roles in host defense mechanisms against heterotypic phage infection, particularly in cluster N mycobacteriophages that infect the host, Mycobacterium smegmatis. Multiple defense systems, encoded by genes expressed from cluster N prophage Butters, specifically target certain phage types, but not others. Several phage genes have orthologues in bacterial species, including several human pathogens. We are investigating these prophage-encoded genes to uncover mechanisms of defense to provide insights into strategies employed by phages and their hosts to counter viral attacks. This work has implications for implementation of phage therapy, for developing phage-resistant bacterial strains for use in industry and biotechnology, and for the use of phage proteins as potential biocontrol agents to mitigate the impact of several bacterial pathogens.
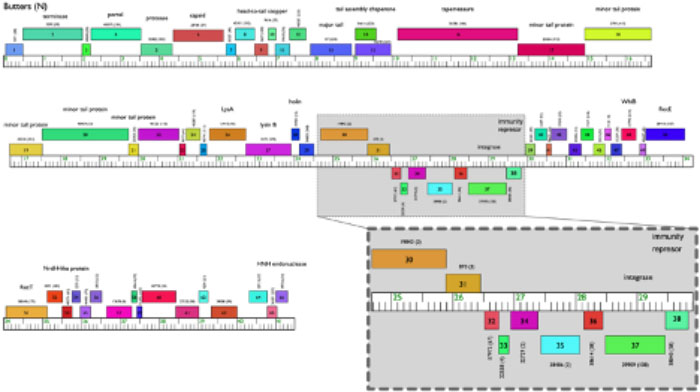
Genome map of mycobacteriophage Butters with predicted functions depicted above the boxes. Genes 30-36 within the grey shadow box are under investigation for their role in prophage-mediated defense against heterotypic viral attack. Map is modified from the Phamerator database Actino_Draft [version 353 (Cresawn SG et al., 2011. BMC Bioinformatics 12: 395)].
Selected ribosomal protein paralogue publications:
Kearse, M.G., Chen, A.S., and Ware, V.C. 2011. Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced. Nucleic Acids Res., 39(7): 2701–2716.
Kearse M, Ireland J, Prem S, Chen A, Ware V. 2013. RpL22e, but not RpL22e-like-PA, is SUMOylated and localizes to the nucleoplasm of Drosophila meiotic spermatocytes. Nucleus 4: 241-258; http://dx.doi.org/10.4161/nucl.25261.
Mageeney CM, Kearse MG, Gershman BW, Pritchard CE, Colquhoun JM, Ware VC. Functional interplay between ribosomal protein paralogues in the eRpL22 family in Drosophila melanogaster (2018). Fly, DOI: 10.1080/19336934.2018.1549419
Mageeney CM, Ware VC. Specialized eRpL22 paralogue-specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster. Molecular Biology of the Cell 12 Jun 2019 https://doi.org/10.1091/mbc.E19-02-0086.
Gershman, BW, Pritchard, CE, Chaney, KP, and Ware, VC. 2020. Tissue-specific Expression of the Ribosomal Protein Paralogue eRpL22-like in Drosophila melanogaster Eye Development. Developmental Dynamics, https://doi.org/10.1002/dvdy.185.
Gershman, BW, Pritchard, CE, Chaney, KP, and Ware, VC. Eye-specific disruption of ribosomal protein paralogue eRpL22-like impacts interommatidial hair cell patterning. Manuscript in preparation.
Selected phage biology publications:
Cresawn, S. G., et al. 2014. Comparative Genomics of Cluster O Mycobacteriophages. PLoS One, Mar 5;10(3):e0118725. doi: 10.1371/journal.pone.0118725.
Pope, W, et al. 2015. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of bacteriophage genetic diversity. eLife;e06416.DOI:10.7554/eLife.06416.
Dedrick, R. M., et al. 2017. Prophage-mediated defence against viral attack and viral counter- defence. Nature Microbiology, 2017 Jan 9;2:16251. doi: 10.1038/nmicrobiol.2016.251.
Mageeney, C. M., et al. 2017. Genome Sequence of Cluster W Mycobacteriophage Taptic, doi: 10.1128/genomeA.01606-16 Genome Announc. March 2017 vol. 5 no. 11 e01606-1.
Mageeney, C. M. et al., 2017. Genome Sequences of Mycobacteriophages Jane and Sneeze, New Members of Cluster G, doi: 10.1128/genomeA.01486-16 Genome Announc. Mar 2017 vol.5 no.11 e01486-1.
Klyczek KK, et al., 2017. Tales of diversity: Genomic and morphological characteristics of forty-six Arthrobacter phages. PLoS ONE 12(7): e0180517. https://doi.org/10.1371/journal.pone.0180517.
Edgington, N., et al., 2017. Genome Sequences of Chancellor, Mitti, and Wintermute, three Subcluster K4 phages isolated using Mycobacterium smegmatis mc2155. Genome Announcements, DOI: 10.1128/genomeA.01070-17.
Hughes, L., et al., 2017. Eight Genome Sequences of Cluster BE1 Phages isolated on Streptomyces. doi: 10.1128/genomeA.01146-17, Genome Announc. January 2018 vol. 6 no. 2 e01146- 17.
Caratenuto RA, III, et al., 2019. Genome sequences of six cluster N mycobacteriophages, Kevin1, Nenae, Parmesanjohn, ShrimpFriedEgg, Smurph, and SpongeBob isolated on M. smegmatis mc2155. Microbiol Resour Announc 8:e00399- 19.https://doi.org/10.1128/MRA.00399-19.
Jacobs-Sera, D., et al., 2020. Genomic diversity of bacteriophages infecting Microbacterium spp. PLoS ONE, 06/2020; 15(6):e0234636.
Mageeney, C.M., et al., 2020. Mycobacterium phage Butters-encoded proteins contribute to host defense against viral attack. mSystems 5:e00534-20.https://doi.org/10.1128/mSystems.00534- 20.
Ware Lab Research overview with undergraduate students
Instruction video: Sucrose Gradient Fractionation