Figure. Overview of Replication Cycle of Herpesviruses
REPLICATION OF dsDNA GENOME VIRUSES
A. Herpesviruses
For the next three weeks, we will be concentrating on the details of how a single virus particle can produce hundreds of copies of itself in a human cell over a time scale of some number of hours. This week, we'll look at two viruses with large double stranded DNA genomes, the Herpesviruses and the Poxviruses.
1. What are the basics of the replication
cycle of Herpesviruses?
Herpesvirus virions are enveloped, with large icosahedral
capsids containing ds DNA genomes (130-230 Kbp) of a hundred or so genes. Between
the icosahedral capsid and the lipid envelope is an amorphous protein layer
called the tegument. Overall, there are about 40 to 50 different proteins in the virion
("virion structural proteins"). Some of these proteins make up the icosahedral capsid, some make up the tegument, and some are the glycoproteins in the envelope.
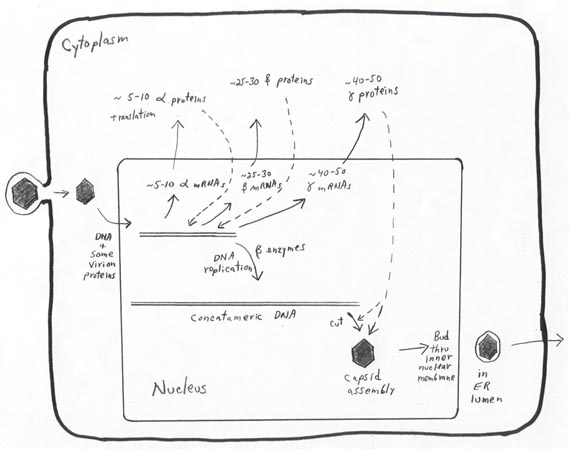
Viral DNA replication, gene expression, and virion assembly are all complex.
An overview and some details of Herpesvirus replication are provided in the "bare-bones" figure below, which pertains most directly to HSV-1 but
is also generally similar for the other herpesviruses. We will add details to
the overview figure as we talk about it.
HSV-1 entry occurs via envelope-plasma membrane fusion at the cell surface. The icosahedral core is transported through the cytoplasm toward the nucleus. At the nuclear pore complex, the viral DNA (with a few DNA-associated proteins) is released through the pore, while the capsid remains intact on the nuclear envelope surface. Once the viral genome is in the nucleus, transcription begins from several "alpha" promoters.
HSV-1 DNA replication occurs via a rolling circle process that produces concatemeric
(i.e. multiple length) copies of the genome. This process is carried out by
enzymes synthesized part way through the infectious cycle (the beta, or "delayed
early" proteins). Later, newly made gamma (late) proteins cut the concatemeric
DNA into unit genome lengths and assemble with the DNA to form new icosahedral
nucleocapsids (in the nucleus).
Expression of the HSV-1 genome involves a three-part temporal pattern of feedback-enhanced
and controlled gene expression (alpha, beta, and gamma) and transport of lots
of newly synthesized viral protein from the site of synthesis (cytoplasm) back
into the nucleus, where virion assembly takes place. Because of this complexity,
figuring out the molecular details of genome replication, gene expression, and
the roles of all of the proteins in the various (~8) human herpesviruses remains
an active area of virology research today.

2. What is our current understanding of how newly assembled
herpesvirus capsids get enveloped and released?
Recent research has shown that the way I have drawn viral envelopment and release
in the figure above is an oversimplification. Studies on cytomegalovirus
( CMV; also called Human Herpes Virus 5, or HHV-5 )
showed that, after envelopment by budding through the inner nuclear membrane,
the particles lose this envelope by budding through the outer nuclear membrane.
The capsids, now in the cytoplasm, get re-enveloped at the Golgi complex, travel
to the cell surface in Golgi-derived vesicles, and get released from the cell
by vesicle fusion with the plasma membrane. All of this is shown in a drawing
in a short "perspectives" article by Sanchez and Spector in Science
(2002).
In 2006, the Journal of Virology published three "letters" on this subject under the overall title "The Egress of Herpesviruses from Cells: the Unanswered Questions." At the end of this posting are links to seven more recent journal articles on this question, from 2007 to 2010.
3. What are some examples of 2010 and 2011 journal articles on herpesvirus
replication?
There is a March 2010 mini-review in the Journal of Virology titled "Early Events in Kaposi's Sarcoma-Associated Herpesvirus Infection of Target Cells".
The January 2011 issue of the Journal of Virology has an article titled "Coordinated Leading and Lagging Strand DNA Synthesis by Using the Herpes Simplex Virus 1 Replication Complex and Minicircle DNA Templates".